Bayesian Statistics in a jiffy!
 Image credit: Unsplash
Image credit: Unsplash
In Statistics, estimating uncertainty of an unknown quantity is a core
problem. Broadly speaking, dealing with this can take one of two paths — Frequentist estimation or Bayesian estimation. It is worth noting, however,
that these two approaches fundamentally have different inference schemes about
how they express degree of belief, and needless to say, there is a
raging 2-century-old debate over what probability means. In this post, I
intend to briefly focus on Bayesian statistics, with demonstrations in R. Basic
knowledge of Statistics is assumed. Further to that, I assume you have, at least,
a basic working familiarity with analysis in R Software.
Let us assume I show you a flip coin and exclaimed; ‘Look! This coin in my hand
is unbias’. But you are not the type of people to accept mere assertions
without overwhelming evidence. So to accept my claim, you demand that we toss
the coin, say,
Referring to the representation above, if we consider prior) before we
even see/observe the data. After looking at the data, our belief of the parameter (estimation)
is updated (posterior). It is because of the subjective nature of the ‘initial belief’ that
makes many frequentist enthusiasts criticize the Bayesian method. However, Bayesian statisticians have refuted this criticism by positing that, in decision-making where uncertainty is present, it’s better to explore all scenarios in a rigorous and coherent manner. I should point out immediately that, these two approaches are not, in reality, contradictory but only presents us an opportunity to address the problem from two different angles.
Template for Bayesian inference
In this section, I will briefly illustrate the so-called ‘prototype’ of Bayesian statistical
inference. Let
where;
prior. The plausible values ofposterior. After seeing the data, what is our updated belief?normalizer. A constant that is of less importance to the modeling process
Example
In a certain population, it is believed that
This is clearly a Bayesian problem. Define
We’re interested in
So what is this value? It represents our updated belief of the proportion of people in the population who are having the disease, thus,
From this simple demonstration, let’s move to a more interesting scenario; a case where
we allow the prior to take more plausible values (rather than a point estimate), observed data,
and the likelihood. I shall demonstrate this in R using ‘brute force’. One way
to do that is to consider
xfun::pkg_attach(c("tidyverse", "glue", "cowplot"))
beta_params <-
tribble(
~ shape1, ~ shape2,
0.5, 0.5,
1.0, 1.0,
5.0, 1.5,
5.0, 5.0,
5.0, 20.0,
50.0, 150.0
)
# Create plot of beta distribution with varying shapes
plot_beta <- function(dat) {
shap1 <- dat[["shape1"]]
shap2 <- dat[["shape2"]]
data.frame(x = c(0, 1)) %>%
ggplot(aes(x)) +
stat_function(
fun = dbeta,
n = 101,
args = list(shape1 = shap1, shape2 = shap2)
) +
ylab("Density") +
xlab("Probability") +
ggtitle(glue("Beta({shap1}, {shap2})")) + theme_grey()
}
beta_plots <-
split(beta_params, seq_len(nrow(beta_params))) %>%
lapply(plot_beta)
plot_grid(
plotlist = beta_plots,
ncol = 3, nrow = 2,
labels = c("I", "II", "III", "IV", "V", "VI"),
align = "none",
label_size = 11
)
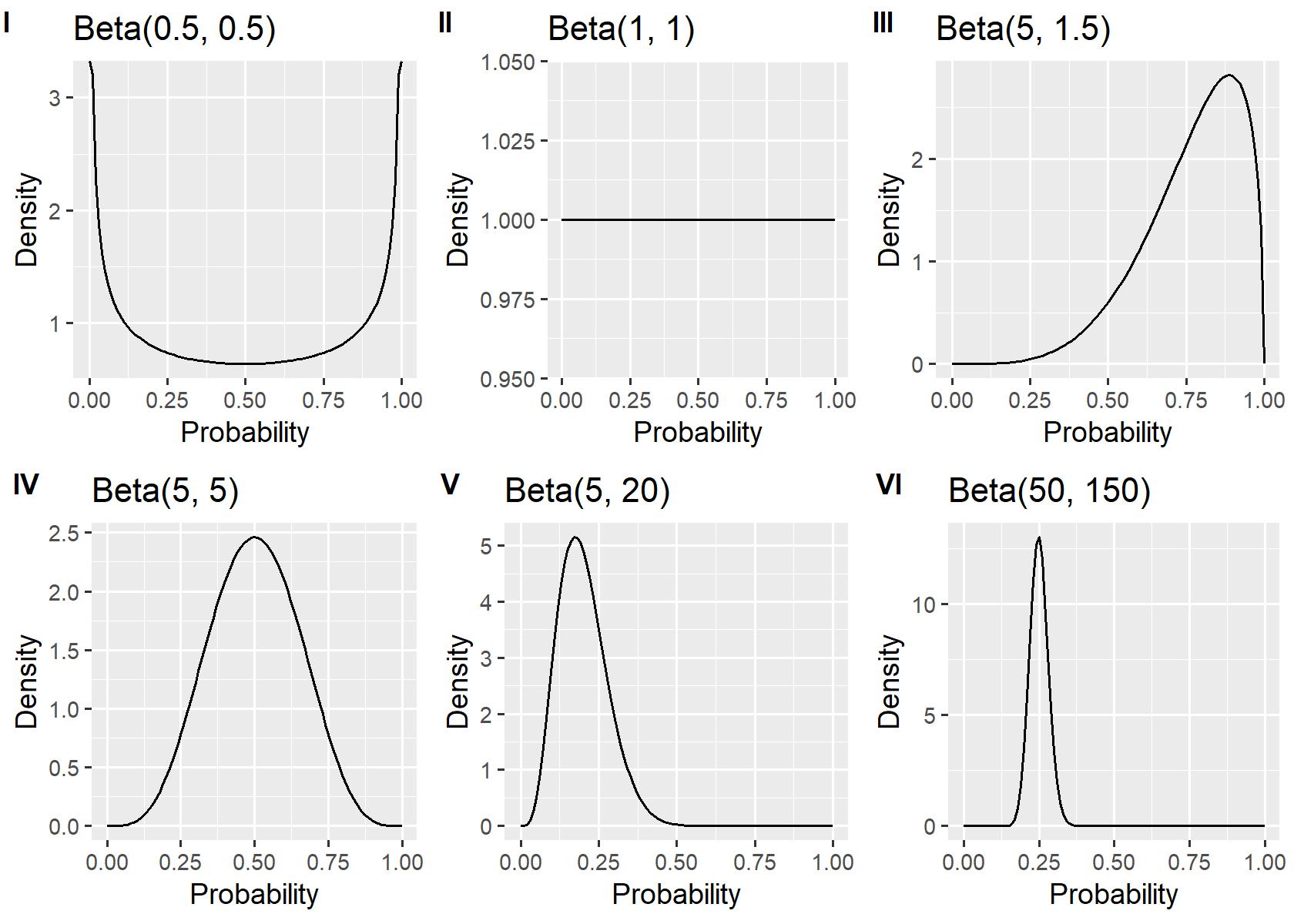
In practice, you could set the prior to any probability distribution of your choice, but some
known probability distributions make the maths easier to handle. For instance,
if we cite the earlier example (about screening a disease X), it is reasonable to think
that the number of people who tested positive for the disease follows a binomial distribution
with an unknown parameter,
Now, consider a hypothetical situation where you decide to check your inbox (email)
for incoming messages. Let’s say you do the checking only between prior to assume various forms, keeping in mind that the number of emails in the
xfun::pkg_attach(c("tidyverse", "reshape2", "glue", "cowplot"))
data = 5
beta_params <-
tribble(
~ shape1, ~ shape2,
0.5, 0.5,
1, 1,
5, 1,
5, 5,
15, 5,
50, 160
)
plot_beta <- function(dat) {
# extract shape1 and shape2 values
cur_shape_1 <- dat[["shape1"]]
cur_shape_2 <- dat[["shape2"]]
param_space <- seq(0, 1, by = 0.001) # parameter space
likelihood <- dbinom(data, size = 20, prob = param_space) # Likelihood
prior <- dbeta(param_space, shape1 = cur_shape_1, shape2 = cur_shape_2) # Prior
weighted_likelihood <- likelihood * prior # numerator
normalizer <- sum(weighted_likelihood) # denominator
posterior <- weighted_likelihood/normalizer # posterior distribution
my_dat <-
melt(data =
tibble(
Theta = param_space,
Likelihood = likelihood * 5,
Prior = prior,
Posterior = posterior*length(param_space)
),
id.vars = "Theta",
variable.name = "Key"
)
ggplot(my_dat, aes(x = Theta, y = value, color = Key)) +
geom_path() +
ylab("Probability density") +
xlab("Parameter space (theta)") +
ggtitle(glue("Beta({cur_shape_1}, {cur_shape_2})"))
}
beta_plots <-
split(beta_params, seq_len(nrow(beta_params))) %>%
lapply(plot_beta)
# Combined plotting
plot_grid(
plotlist = beta_plots,
ncol = 2, nrow = 3,
labels = c("I", "II", "III", "IV", "V", "VI"),
align = "none",
label_size = 11
)
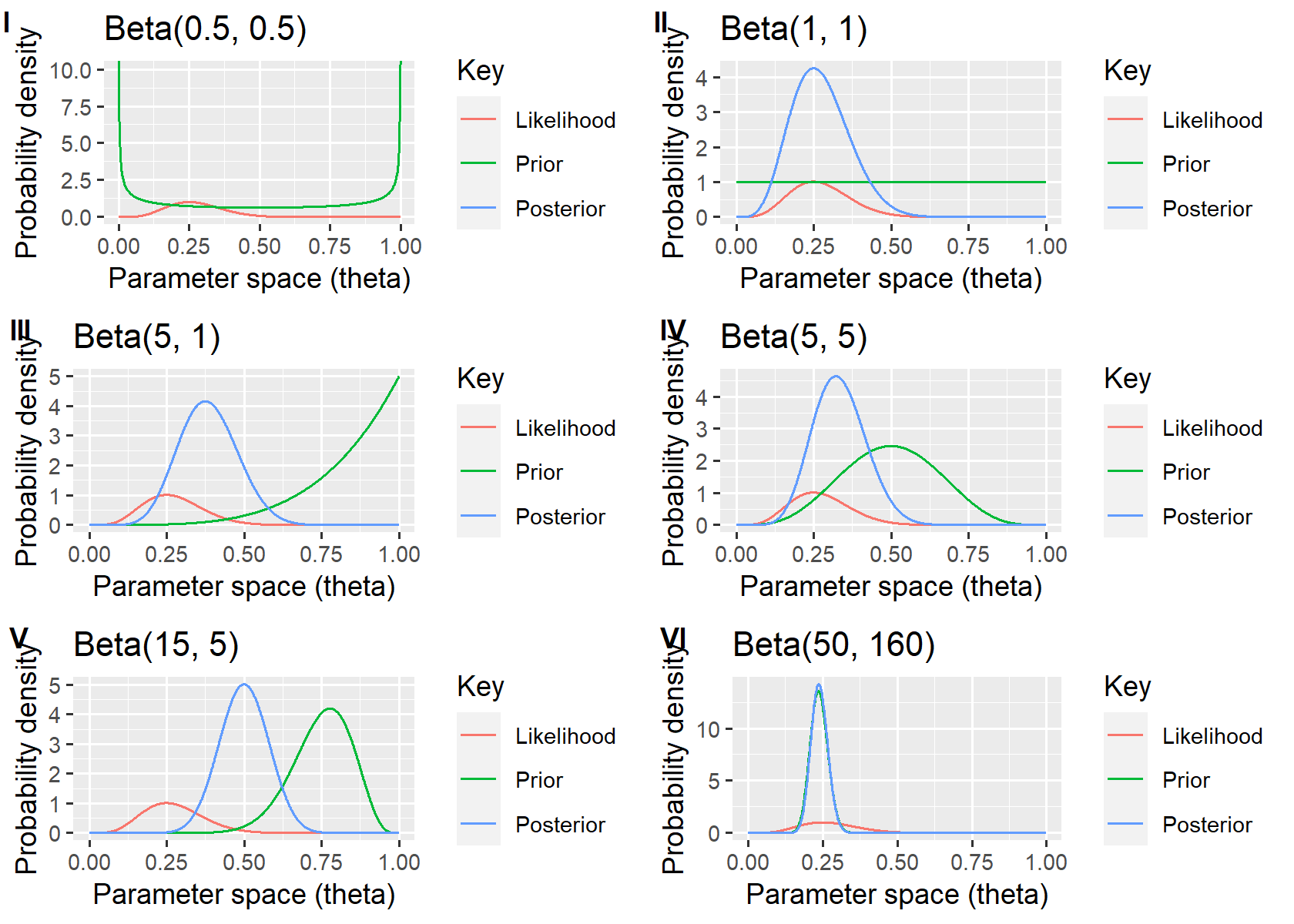
From the figure, taking for example
Beta(1,1) is the same as the Unif(0, 1), uniform distribution.
Concluding remarks
In this post, I have briefly touched on Bayesian Statistics – where we start the estimation process with an initial ‘belief’, and as the modeling processes continue, we get to update the so-called belief. Thus, before we look at the data, what possible is the value of the parameter, non-informative prior as against informative priors which generally incorporate external information into the modeling process. For instance, in our ‘email experiment’, we dealt with one trial, relying on ‘non-informative prior’. To go deeper, we could have decided to collect more data, by way of repeating the trials several times, and investigating the behavior of the estimation for a prior, for instance,